
Multicellular organisms are composed of diverse cell types that originate from a single cell. Cellular identities are orchestrated in part through action of key factors on regulatory elements. In diploid organisms, complexity of regulatory interactions, which influence gene expression, is increased due to the presence of homologous maternal and paternal chromosomes. However, as much of our current view of cell diversity is derived from genome organization and regulation of either single cells or groups of cells, much less is known how parental genomes contribute to achieve this diversity. Moreover, it is unclear how this diversity evolutionarily relates to health and disease states. To this end, we deploy haplotype-specific regulatory genomics, bioinformatics, quantitative imaging, and genetics.
Single-cell chromatin dynamics during transcriptional switches
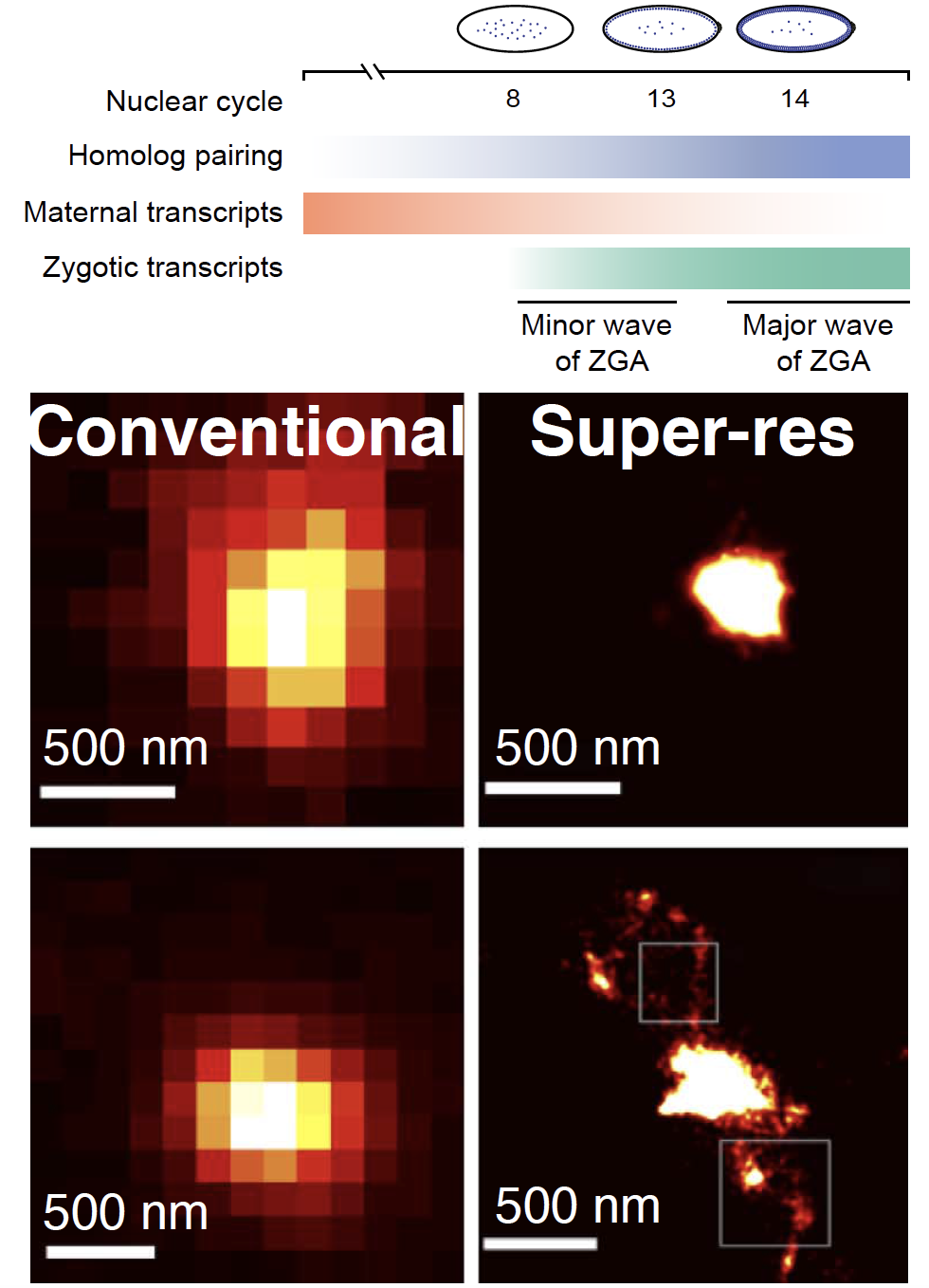
To achieve cellular identities dynamic and fine-tuned spatio-temporal regulation is essential when multipotent cells become specified into different lineages. We investigate chromatin dynamics during a crucial period as the embryonic genome awakens. We employ single-cell Oligopaints-based FISH technology at homolog-specific and multiplexed single-molecule super-resolution level.
3D regulatory genome in the light of evolution and disease
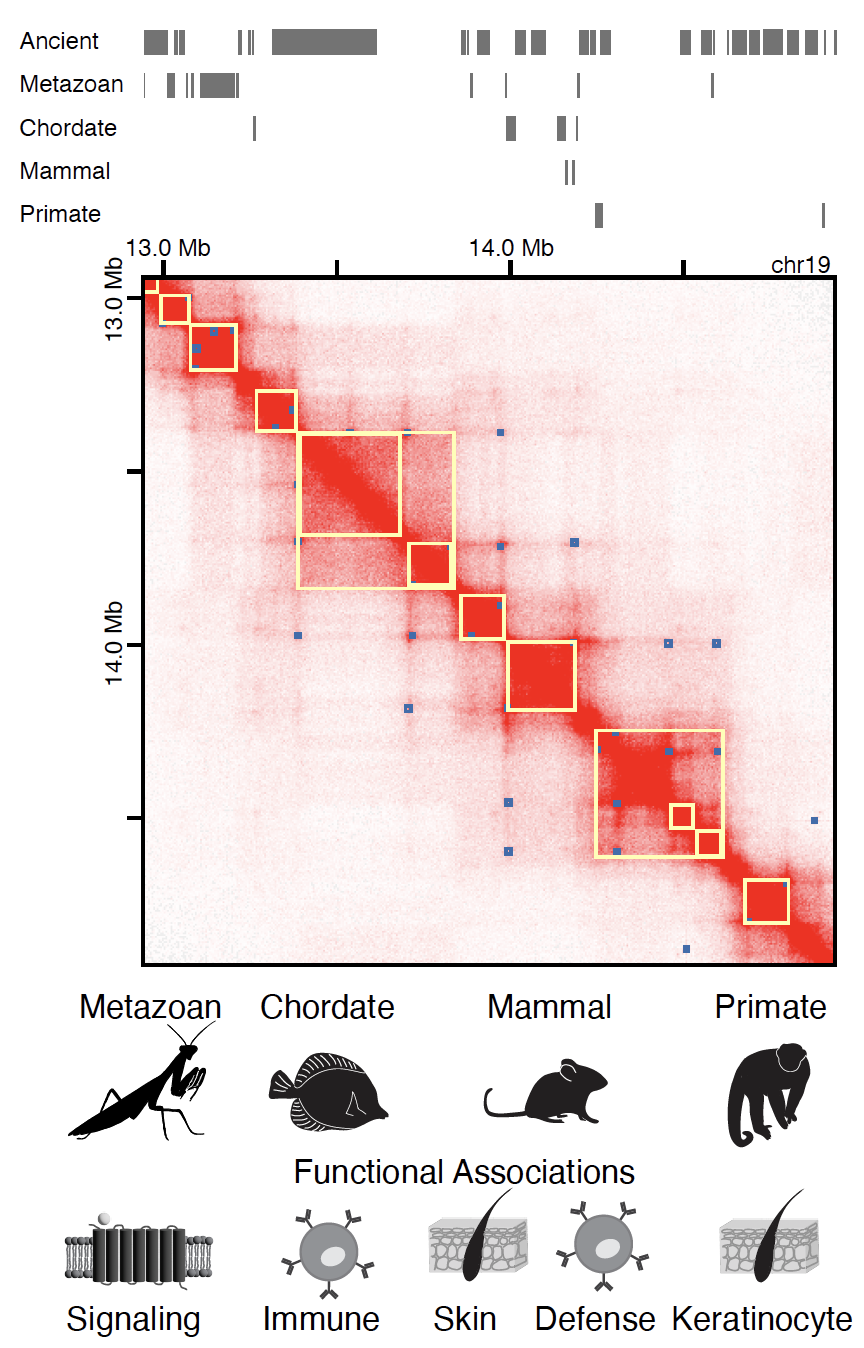
We also leverage our approaches from Drosophila to mammalian systems to investigate conservation behind genome folding and regulation. Revealing relationships between gene evolution and 3D genome organization in healthy and cancer states can present a major step forward in our understanding of the tissue-specific regulatory programs in development and disease.
Distribution of introns across the 3D genome and their functional significance
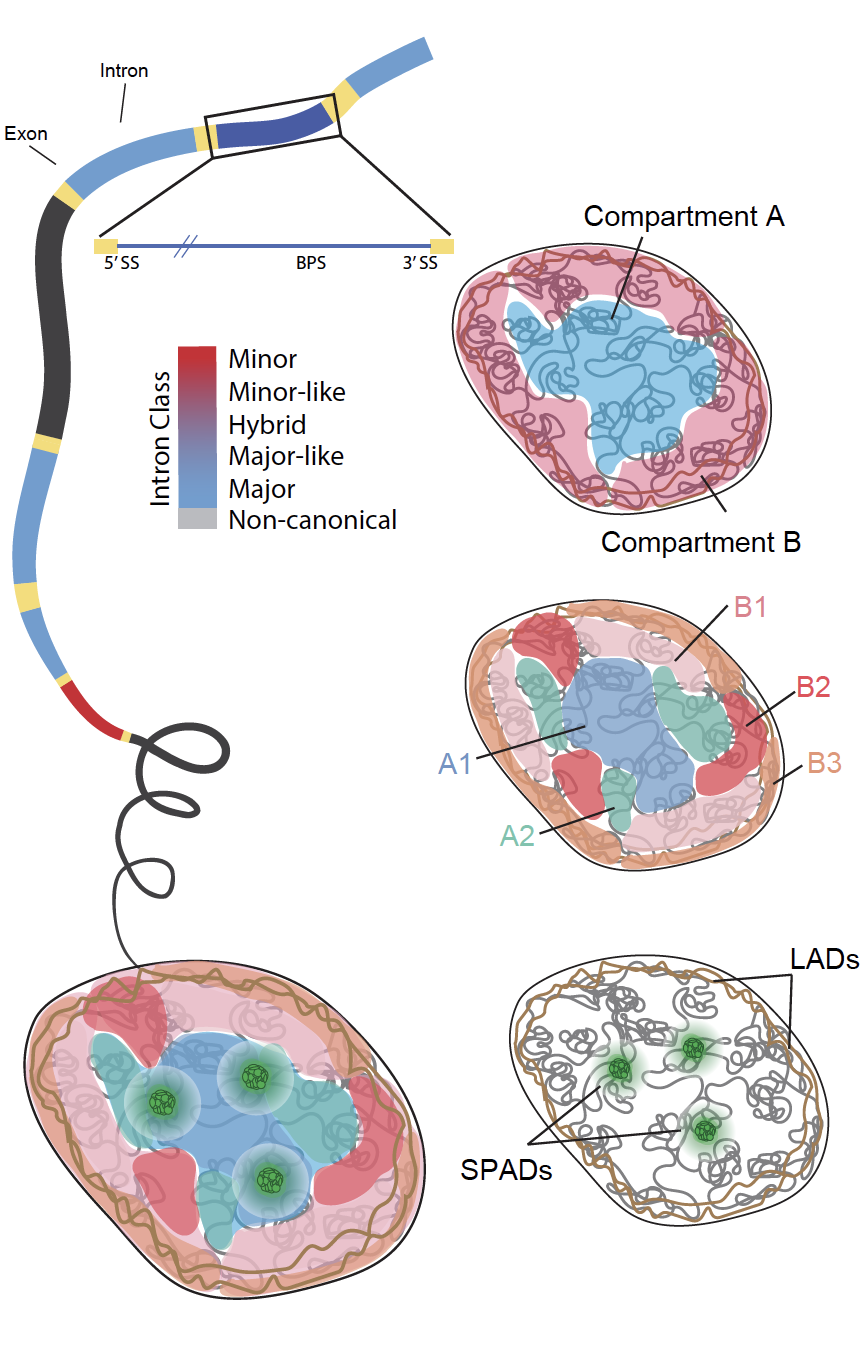
Introns occupy 98% of the protein coding genome and have an often-overlooked influence on the regulation and expression of eukaryotic genes. We examine spatial organization and splicing of intron classes in the human genome to uncover how intron organization relates to the functional regulation of the genome, contributes to cell-type diversity, and changes with disease.